DrugnomeAI Framework
DrugnomeAI is an adapatation of Mantis-ML that provides both disease-agnostic and disease-specific gene druggability framework, implementing stochastic semi-supervised learning on top of scikit-learn and keras/tensorflow. Drugnome takes its name from a contraction of the 'druggable genome'.
Citation:
DrugnomeAI is an ensemble machine-learning framework for predicting druggability of candidate drug targets
Arwa Raies, Ewa Tulodziecka, James Stainer, Lawrence Middleton, Ryan S. Dhindsa, Pamela Hill, Ola Engkvist, Andrew R. Harper, Slavé Petrovski & Dimitrios Vitsios✉
Communications Biology, 5, 1291 (2022)
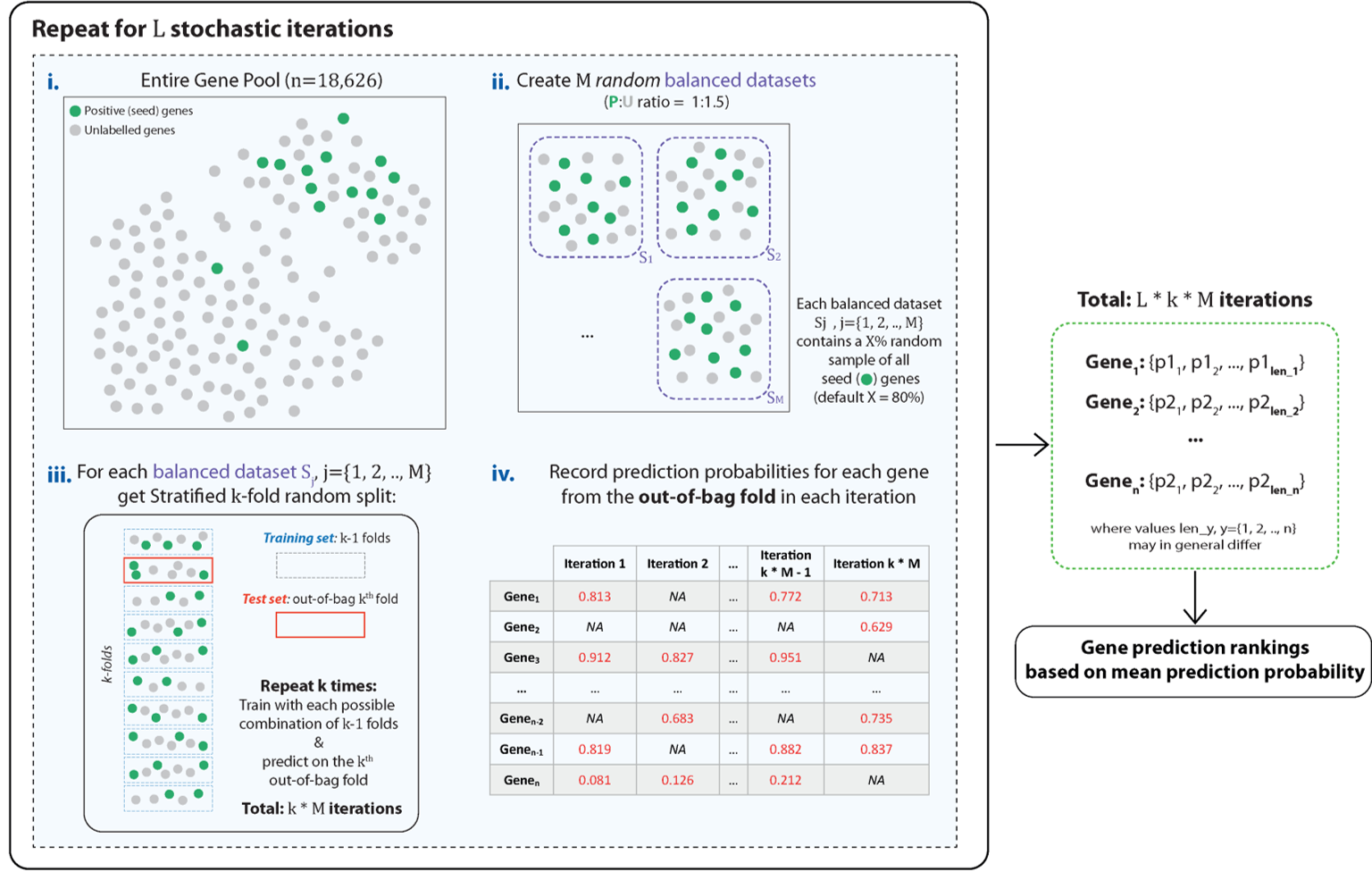
Mantis-ML is a pre-existing ML framework that was built in order to generate novel gene-disease associations.
The original framework automatically performs pre-processing steps, such as data cleaning and exploratory data analysis, then performs learning and classification.
In brief, the learning algorithm classifies genes as either 'druggable' or 'not-druggable' based on a subset of known examples.
Typically, the number of known positive examples is far outweighed by the number of negative examples, inducing a class imblance problem.
- The imbalanced data sets are segregated into balanced data sets (sampling unlabelled genes as negative examples).
- Then for each balanced data set k-fold cross validation is performed, creating a prediction on the k'th fold at each step.
- The probabilities of each gene being classified are recorded.
- Finally, a consensus across all predictions is generated, based on their mean prediction probability.
Drugability definition & labelling
For our ML framework to train on the data, we were first required to define what ‘druggable’ means. We describe druggability in DrugnomeAI with two different sets of labels, these being Pharos labels and tier labels.

Tclin are approved drug targets
Tchem have small molecule activities in ChEMBL

Tier 1 genes include targets of approved small molecules and biotherapeutic drugs (as well as clinical-phase drug candidates)
Tier 2 genes encode targets with known bioactive drug-like small-molecule binding partners as well as those with high sequence similarity with approved drug targets
Tier 3 genes encode secreted or extracellular proteins that have only distant similarity to approved drug targets,
as well as other members of key druggable gene families not already included in Tier 1 or 2
For the purpose of custom DrugnomeAI models, we leveraged three types of drug modalities: small molecule, antibody, and PROTAC, for which we trained models on genes targeted by small molecules, antibody, and PROTACs, respectively. In this case, target genes have been specified based on retrieved genes lists from the Open Targets platform. In the following section, you can find descriptions of the target groups for all the specialized models we developed.
Datasets for specialized models
Modality specific
Oncology/non-oncology
Additional druggability evidence
Provided alongside the derived DrugnomeAI druggability scores are additional gene annotations and properties related to their druggability profile. Gene-specific evidence includes:
- Modalities Two tractability modalities, one for antibody (AB) and one for small molecule (SM). These are not necessarily approved drugs - they could be in clinical trials for example - and additional distinction is provided between Approved drug, Advanced clinical, and Phase 1 clinical, with everything else labelled as Experimental.
- Cellular localisation Cellular localisation indicating any of Intracellular, Membrane or Secreted. Original data derived from UniProt.
- Orthologs in other species Gene homologs across different species.
- Gene and protein expression Gene and protein expressions quantify the extent of the expression in different tissues (organised into either organs or anatomical systems). The resource compiles data from Expression Atlas, Human Protein Atlas and Genotype-Tissue Expression (GTEx) Program. The data presented here was extracted using the OpenTargets API, exposing expression levels organised into EFO codes, label, organ and anatomical system. Where multiple entries existed for a given organ or anatomical system (for example differing only in their EFO code), expression levels were aggregated across the finer categorisation. For RNA expression, the aggregation function was taken to be the mean and for protein expression the aggregation function was taken to be the maximal value.